Archival Notice
This is an archive page that is no longer being updated. It may contain outdated information and links may no longer function as originally intended.
Home | Glossary | Resources | Help | Contact Us | Course Map
Features have been added in GeneMapper® ID Software v3.2 to facilitate analysis of the AmpF?STR® Yfiler™ PCR Amplification Kit.
Features and Procedures (v3.2) | |
|---|---|
| Feature/Procedure | Description |
| New feature: Allele calling parameters for new marker repeat types | In GeneMapper ID Software v3.1, allele-calling parameters were only available for markers with tetranucleotide repeat motifs. In GeneMapper® ID Software v3.2, however, allele calling parameters are available for four marker repeat types: tri-, tetra-, penta-, and hexanucleotide. The four allele calling parameters appear in the Allele tab of the Analysis Method Editor. All related analysis values are entered directly into the Tri, Tetra, Penta, or Hexa column fields, which allows for viewing of all values. |
| New feature: Plus stutter filtering (for the DYS392 locus) | To aid in interpreting genotype profiles, two new fields have been added to the Allele tab of the Analysis Method Editor specifically to filter out the DYS392 plus stutter:
|
| Procedure: Workaround for the DYS19 locus (-2bp filtering) | Laboratories may choose to implement a work-around for the DYS19 locus by using the Minus A Ratio and Minus A Distance fields in the Allele tab of the Analysis Method Editor. These fields can be used to filter out the -2-bp stutter that is observed at the DYS19 locus. |
| Procedure: Creating HID analysis methods for the Yfiler kit | Use the Analysis Method Editor in GeneMapper® ID Software v3.2 to set analysis parameter values for analyzing the Yfiler kit data. |
| Procedure: Creating a table setting and uploading exported haplotype(s) for searching profiles with the Yfiler™ Haplotype Database | Using the Table Setting Editor, a table setting is created in GeneMapper® ID Sortware v3.2 to export haplotypes specifically for searching the Yfiler Haplotype Database for profile match estimation. |
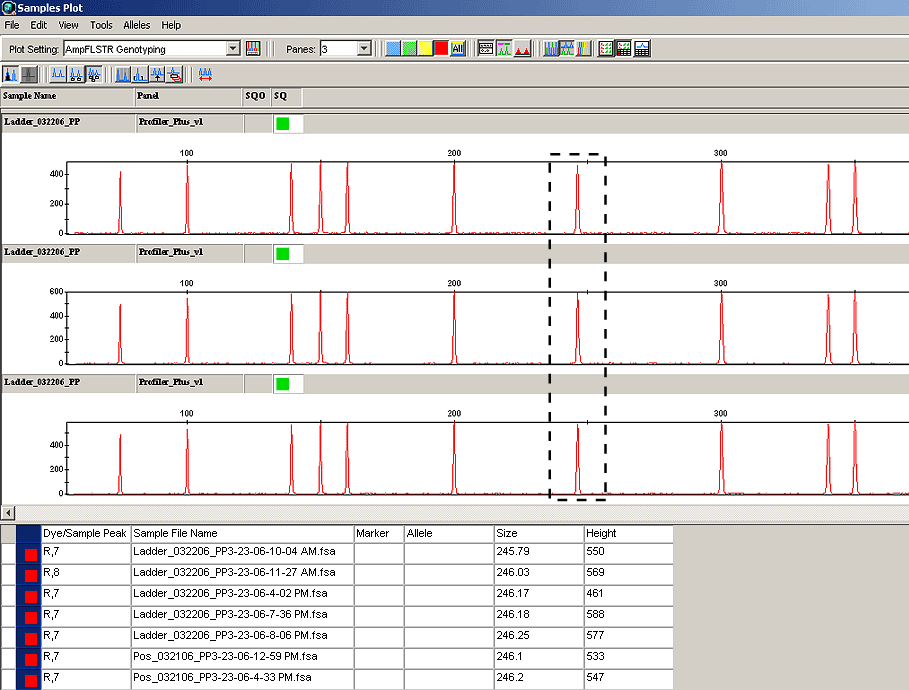
Genotyper® had a feature that allowed review of the sizing standard for precision assessment. This feature was not available in the first version of GMID (v3.1); it is included in version 3.2. However, it requires a work-around to achieve the precision statistics on the 250 base pair fragment. The size standards can be overlayed from all samples in the run. The base pair sizes for the fragments can be displayed, the table exported to Microsoft Excel, and the Excel functions used to calculate the 250 base pair precision from a run.
View a screen capture of the size standard overlay with the sizing table beneath:
Additional Online Courses
- What Every First Responding Officer Should Know About DNA Evidence
- Collecting DNA Evidence at Property Crime Scenes
- DNA – A Prosecutor’s Practice Notebook
- Crime Scene and DNA Basics
- Laboratory Safety Programs
- DNA Amplification
- Population Genetics and Statistics
- Non-STR DNA Markers: SNPs, Y-STRs, LCN and mtDNA
- Firearms Examiner Training
- Forensic DNA Education for Law Enforcement Decisionmakers
- What Every Investigator and Evidence Technician Should Know About DNA Evidence
- Principles of Forensic DNA for Officers of the Court
- Law 101: Legal Guide for the Forensic Expert
- Laboratory Orientation and Testing of Body Fluids and Tissues
- DNA Extraction and Quantitation
- STR Data Analysis and Interpretation
- Communication Skills, Report Writing, and Courtroom Testimony
- Español for Law Enforcement
- Amplified DNA Product Separation for Forensic Analysts