Archival Notice
This is an archive page that is no longer being updated. It may contain outdated information and links may no longer function as originally intended.
Home | Glossary | Resources | Help | Contact Us | Course Map
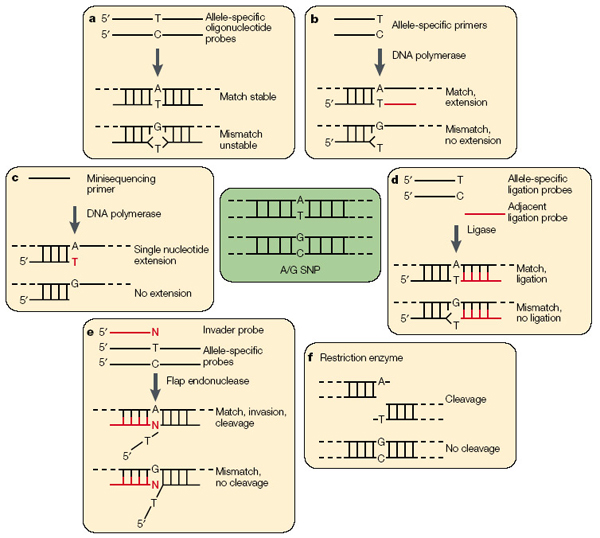
Spurred on by the need to type thousands of SNPs (Single Nucleotide Polymorphisms) for human disease association studies, a variety of technologies have been developed for SNP analysis. Most methods are amplification based and the SNP is subsequently detected by primer extension, oligonucleotide ligation, or hybridization of a probe to the amplified product (see figure below). In some ways, the best SNP-typing system is that of standard DNA Sanger sequencing, in which the SNP site and flanking sequences are simultaneously interrogated. However, such an approach is limited to densely packed SNPs, such as those present in the mtDNA control region. For the other SNP applications in which the SNP sites are separated by thousands (often hundreds of thousands) of bases, Sanger sequencing is neither practical nor cost effective.
The SNP analysis technology that has been used most productively for the coding region mtDNA, physical characteristics, and degraded DNA applications is based upon primer extension (or mini-sequencing) followed by capillary electrophoresis (CE) or solid phase capture.09 The SNaP shot™ assay (AB), which has been used for the mtDNA coding region analysis, uses a primer that abuts the SNP site, genomic DNA template, DNA polymerase, and fluorescently labeled terminator ddNTPs. The SNP site on the genomic template will determine, by complementary Watson-Crick base pairing, which ddNTP will be extended and incorporated into the primer. The now fluorescently labeled extended primer is separated by capillary electrophoresis, and the allelic status determined by the nature of the fluorescent dye detected. Multiplexing is achieved by designing primers of different lengths (or containing mobility modifiers) at the primers' 5' ends, such that each separate SNP in the multiplex has a characteristic mobility.
Read more on the SNap shot™ assay on Applied Biosystems website.
The SNPstream® UHT (Orchid Cellmark) is a primer extension methodology that has been used to predict bio-geographical ancestry and to analyze degraded DNA. This platform uses SNP extension primers that are modified at the 5' ends with oligonucleotide tags that are complementary to oligonucleotides that are bound to a solid microplate support. Each well of the microplate has 16 anti-tag sequences. Hybridization to it of a SNP primer extension reaction, previously carried out in solution, permits multiplex typing by location (determines the SNP locus) and bound dye (determines the allelic status).
SNP Analysis Case Example |
|---|
| The investigation of a series of five unsolved serial murders in southern Louisiana between September 2001 and March 2003 was aided by the use of AIM-SNPs. Prior to their use, psychological profiling had indicated the likelihood that a Caucasian male was the culprit. However, AIM-SNP analysis revealed that the killer was likely to be of Black ancestry. Acting upon this lead, investigators eventually arrested an Black suspect, Derek Todd Lee and tried him for the murder of Charlotte Murray Pace. Lee was subsequently linked by DNA evidence to seven other homicides from 1998 to 2003. |
Additional Online Courses
- What Every First Responding Officer Should Know About DNA Evidence
- Collecting DNA Evidence at Property Crime Scenes
- DNA – A Prosecutor’s Practice Notebook
- Crime Scene and DNA Basics
- Laboratory Safety Programs
- DNA Amplification
- Population Genetics and Statistics
- Non-STR DNA Markers: SNPs, Y-STRs, LCN and mtDNA
- Firearms Examiner Training
- Forensic DNA Education for Law Enforcement Decisionmakers
- What Every Investigator and Evidence Technician Should Know About DNA Evidence
- Principles of Forensic DNA for Officers of the Court
- Law 101: Legal Guide for the Forensic Expert
- Laboratory Orientation and Testing of Body Fluids and Tissues
- DNA Extraction and Quantitation
- STR Data Analysis and Interpretation
- Communication Skills, Report Writing, and Courtroom Testimony
- Español for Law Enforcement
- Amplified DNA Product Separation for Forensic Analysts