Archival Notice
This is an archive page that is no longer being updated. It may contain outdated information and links may no longer function as originally intended.
Home | Glossary | Resources | Help | Contact Us | Course Map
Mitochondrial DNA (mtDNA) sequencing technology is routinely used by a limited number of forensic laboratories worldwide because direct sequencing is time consuming and labor intensive.
Direct sequencing determines the order of bases along the template DNA. Automated fluorescent sequence technology has significantly increased the throughput of sequence information in the scientific communities. Often small quantity and degraded DNA are the only samples available in mtDNA forensic casework. The DNA is amplified and directly sequenced using the Sanger method.
The amplified product is:27,30
- Digested with SAP-EXO I (shrimp alkaline phosphatase and exonuclease I) to remove excess deoxynucleotide triphosphate (dNTPs) and PCR (Polymerase Chain Reaction) primers
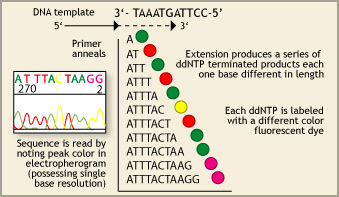
- Exposed to a sequencing primer and incorporation of dideoxyribonucleotide triphosphate (ddNTPs) for the sequencing reaction
- Denatured
- Separated by electrophoresis
Each ddNTP is labeled with a different fluorescent dye. Once the ddNTP is incorporated, chain elongation is terminated. The sequencing primers direct the sequencing of the forward or reverse reaction for each of the DNA strands. Interpretation of the data can be laborious, even with sophisticated software programs.
The amplification and sequencing primer sets are not available commercially in kits similar to the STR (Short Tandem Repeat) PCR amplification kits; however, these primers are published and can be ordered through commercial vendors.31,32 Highly degraded DNA may not amplify efficiently with the primer sets for HV1 and HV2. Miniprimer sets have been developed to amplify smaller portions of HV1 and HV2 and have been successful in overcoming failure to amplify with the larger primers.33
Computer software programs, such as Sequencher™ (Gene Codes), are available to aid the analyst in the tedious task of data analysis.
Sequencher™ is a sequence analysis software program that:
- Builds projects for each sample
- Removes any sequence data of the primers
- Removes poor sequence data based on signal and spacing
- Produces complementary data of the reverse sequence reactions
- Aligns forward and reverse sequencing reactions
- Compares the sequence to the rCRS
A forensic DNA analyst independently examines, interprets, and edits sequencing results prior to reporting.
Additional Online Courses
- What Every First Responding Officer Should Know About DNA Evidence
- Collecting DNA Evidence at Property Crime Scenes
- DNA – A Prosecutor’s Practice Notebook
- Crime Scene and DNA Basics
- Laboratory Safety Programs
- DNA Amplification
- Population Genetics and Statistics
- Non-STR DNA Markers: SNPs, Y-STRs, LCN and mtDNA
- Firearms Examiner Training
- Forensic DNA Education for Law Enforcement Decisionmakers
- What Every Investigator and Evidence Technician Should Know About DNA Evidence
- Principles of Forensic DNA for Officers of the Court
- Law 101: Legal Guide for the Forensic Expert
- Laboratory Orientation and Testing of Body Fluids and Tissues
- DNA Extraction and Quantitation
- STR Data Analysis and Interpretation
- Communication Skills, Report Writing, and Courtroom Testimony
- Español for Law Enforcement
- Amplified DNA Product Separation for Forensic Analysts