Archival Notice
This is an archive page that is no longer being updated. It may contain outdated information and links may no longer function as originally intended.
Home | Glossary | Resources | Help | Contact Us | Course Map
Forensic Applications
Mitochondrial DNA (mtDNA) analysis is an appropriate method for:
- Charred remains
- Degraded specimens
- Old skeletal and fingernail samples
- Hair shafts
These samples either do not contain nuclear DNA or will have experienced a marked reduction in the quality and quantity of nDNA (nuclear DNA) present. In contrast, they contain much larger amounts of mtDNA and sufficient material for typing will remain even after degradation caused by environmental challenges or the passage of time.08
Additionally, mtDNA is quite useful in forensic investigations of remains recovered of a missing person or mass disaster. These remains are often highly fragmented or only very small sample sizes are recovered, such as a single tooth or a sliver of a bone.09 Biological material from known, maternal relatives, even quite distant, can be used as a reference for direct comparison to the recovered remains.
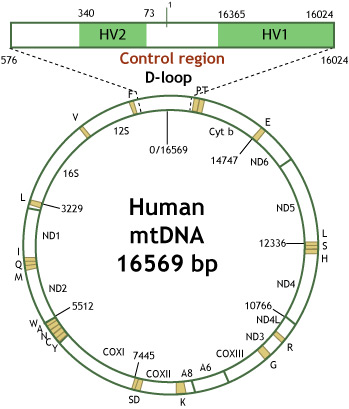
The D-loop contains two regions within its 1.1 Kb fragment that demonstrate multiple variations between individuals. The two variable regions, hypervariable region 1 (HV1) and hypervariable region 2 (HV2) are amplified, detected, and analyzed for forensic identification. Other regions within the mtGenome have been successfully analyzed but are not the typical regions tested for casework.10,11
The origin of replication is between HV1 and HV2. Counting of the bases, or the numerical address along the mtGenome, is initiated at the origin of replication on the heavy strand starting at position 1 (p1) to p16,569. As mentioned earlier, the most common practice for forensic laboratories is reporting sequence information is HV1 and HV2. The sequence information covered for HV1 is p16,024 to p16,365. Likewise, the sequence information covered for HV2 is p73 to p340.
See the YouTube Terms of Service and Google Privacy Policy
Unlike STR (Short Tandem Repeat) analysis where discrete alleles according to size are reported, mtDNA analysis reports the observed base sequence. The standard for the forensic community is to report the sequence information as compared to the revised Cambridge Reference Sequence's (rCRS) light strand, also referred to as the Anderson sequence.12 When the base sequence is the same, then no reference to that particular position is noted. However, if the base sequence at a position is different at one or more positions when compared to the rCRS, then the difference(s) is noted. For a common example of a transition in HV1, the rCRS is an A at p73 and the sequenced sample is a G at p73; a report is generated showing an A to G transition a p73.
View the Cambridge Reference Sequence
In Caucasians, the most common mtDNA types from HV1 and HV2 occur in approximately 7% of the general population. On average, there are eight nucleotide differences between individuals amongst unrelated Caucasians and 15 nucleotide differences between individuals amongst unrelated Africans.13,14
Other Applications
Scientists have used mtDNA analysis for medical studies, evolutionary studies, migration studies, genealogical studies, and historical identifications.
Additional Online Courses
- What Every First Responding Officer Should Know About DNA Evidence
- Collecting DNA Evidence at Property Crime Scenes
- DNA – A Prosecutor’s Practice Notebook
- Crime Scene and DNA Basics
- Laboratory Safety Programs
- DNA Amplification
- Population Genetics and Statistics
- Non-STR DNA Markers: SNPs, Y-STRs, LCN and mtDNA
- Firearms Examiner Training
- Forensic DNA Education for Law Enforcement Decisionmakers
- What Every Investigator and Evidence Technician Should Know About DNA Evidence
- Principles of Forensic DNA for Officers of the Court
- Law 101: Legal Guide for the Forensic Expert
- Laboratory Orientation and Testing of Body Fluids and Tissues
- DNA Extraction and Quantitation
- STR Data Analysis and Interpretation
- Communication Skills, Report Writing, and Courtroom Testimony
- Español for Law Enforcement
- Amplified DNA Product Separation for Forensic Analysts