Crime scenes, particularly those involving violence, often yield evidence from bodily fluids – blood, saliva, urine, semen – left behind. However, discerning the composition and source of the fluids is more complicated than one might think, particularly when several sources are mixed together. If forensic scientists can identify a fluid’s source, they might gain critical information about a person’s involvement with a crime and what happened. In the tragic case of five-year-old Harmony Montgomery, whose body has still not been recovered, investigators were able to reconstruct events by analyzing fluid-soaked ceiling tiles retrieved from a shelter the Montgomery family had visited. They found blood evidence that led prosecutors to allege Montgomery’s father had temporarily stored her body in the shelter’s ceiling before he later moved it.[1]
In recent years, forensic investigators expressed the need for body fluid identification technologies that identify fluids more accurately and with greater sensitivity than traditional immunoassay methods.[2] Fortunately, several new methods have been introduced that meet this demand. Collectively referred to as “omic”-based methods, they are now available for fluid identification purposes (Table 1). National Institute of Justice (NIJ)-funded researchers recently released the results of a study designed to compare the specificity and sensitivity of these “omic”-based methods.
Table 1. Emerging “omic”-based approaches to body fluid identification, including epigenomics, transcriptomics, and proteomics.
| Field | Body Fluid Identification Technology | How the Technology Works |
|---|---|---|
| Epigenomics | DNA methylation analysis | Provides information about the biological source of DNA using DNA methylation patterns (adding methyl groups to DNA, possibly altering gene expression patterns). DNA methylation analysis generally distinguishes between blood, saliva, semen, and vaginal secretions. |
| Transcriptomics | mRNA profiling | Different body fluid types contain different mRNA species. mRNA profiling technology identifies fluid type by analyzing gene expression patterns from the most abundant types of mRNA present. mRNA profiling can detect multiple fluids in a single reaction, including blood, saliva, semen, menstrual blood, and cervicovaginal fluid. |
| Proteomics | Shotgun proteomics and targeted proteomics | Shotgun and targeted proteomics identify body fluids by extracting the protein from the fluid, breaking the proteins down into smaller peptides, separating the peptides using liquid chromatography, and finally identifying the peptides using mass spectrometry. The resulting data are used to identify proteins based on their characteristic peptide fingerprint. Shotgun proteomics is akin to large-scale blind screening of samples, while targeted proteomics is the focused analysis of a pre-defined set of proteins of interest. Shotgun and targeted proteomics can be used to identify serum, plasma, urine, stool, saliva, tears, sweat, and cerebrospinal fluid. |
Dr. Mirna Ghemwari, from the Center for Forensic Science Research and Education, combined expertise with Dr. Jack Ballantyne (transcriptomics), Dr. Bruce McCord (epigenomics), Dr. Glendon Parker (shotgun proteomics), and the New York Office of the Chief Medical Examiner (targeted proteomics). They investigated the specificity and sensitivity of epigenomic, transcriptomic, and proteomic methods for body fluid identification, as well as their accompanying error rates, so crime labs could make informed decisions about which approach to use.
Procedure Standardization and Sample Testing: A Two-Phase Approach
In the first phase, researchers standardized sample preparation methods to ensure fluid technology calibration. Ghemrawi sent samples from five fluid types (semen, venous blood, saliva, vaginal fluid, and menstrual blood) to external labs for identification. The source of the body fluids was withheld from the testing labs until all results were gathered and compared. Analyses indicated that the best results were generated when samples were not diluted with water and swab slurries were not used.
In the second phase, researchers analyzed mock casework samples. They deposited 58 samples consisting of nine fluid types[3] on different types of substrates at a variety of volumes. Then, they identified the fluid type using six methods: two types of traditional immunoassay (methods A and B), DNA methylation, mRNA testing, shotgun proteomics, and targeted proteomics (Figure 1).
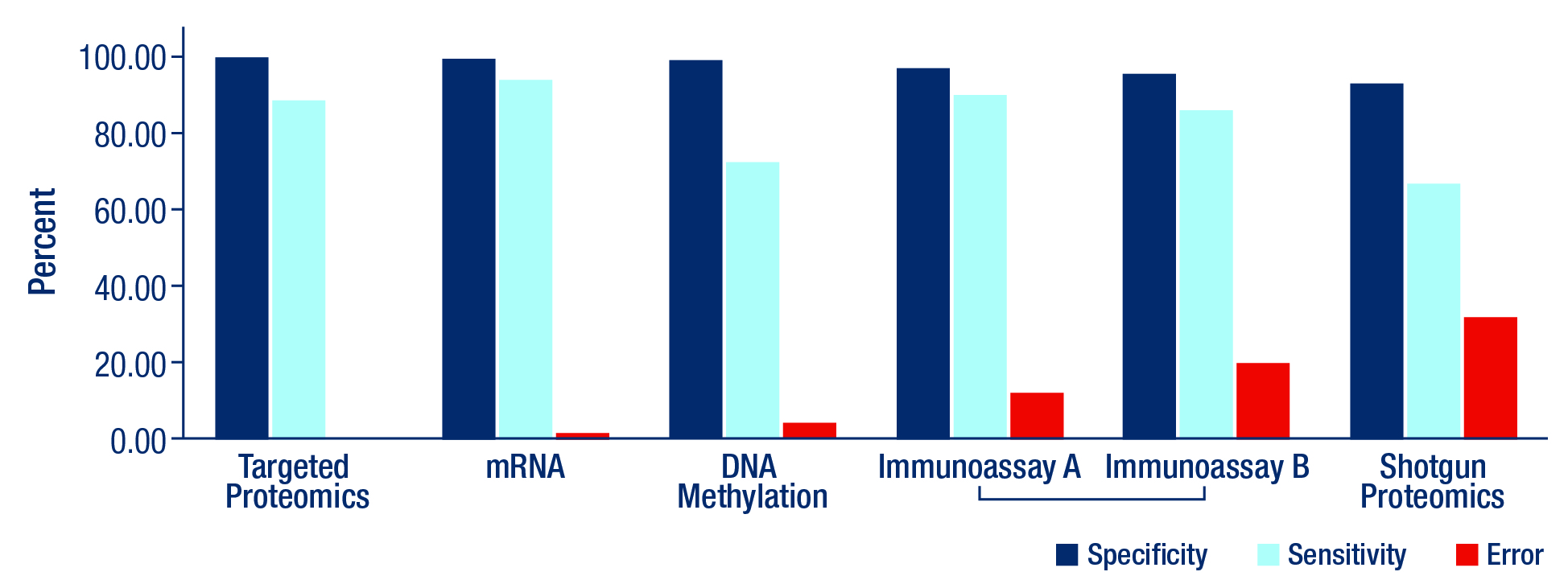
Figure 1. Comparison of specificity, sensitivity, and error rates of the body fluid identification assays: targeted proteomics, mRNA, DNA methylation, the traditional immunoassay methods A and B[ii], and shotgun proteomics.
Researchers evaluated the performance of each method and found:
- Targeted proteomics and mRNA assays performed best across all three metrics with the highest specificity, the highest sensitivity, and the lowest error rates.
- The specificity rates were highest for targeted proteomics (100%), mRNA assays (99.7%), and DNA methylation (99.5%), while the specificity rates for the traditional immunoassay methods (96%, averaged) and shotgun proteomics (93.4%) were lower.
- The sensitivity rates were highest for mRNA assays (94.1%), targeted proteomics (88.5%), and immunoassays (87.1%, averaged), while the sensitivity rates for DNA methylation (72.5%) and shotgun proteomics (67.7%) were lower.
- The error rates were lowest for targeted proteomics (0%), mRNA assays (1.5%), and DNA methylation (3.8%), while the error rates were higher for the immunoassays (15.9%, averaged) and shotgun proteomics (32.3%).
Notably, the two proteomic techniques (targeted and shotgun) had strikingly different error rates. The NY Office of the Chief Medical Examiner achieved no false positives using the targeted proteomics technique, whereas shotgun proteomics had the highest error rate of all the assays. The screening foci of these proteomic techniques could account for the error rate difference, since targeted proteomics uses a much smaller, pre-defined and well-characterized set of proteins of interest it is therefore less likely to misidentify a fluid.
Advancing Criminal Justice
Proper body fluid identification from crime scenes could lead to more accurate analyses of evidence and aid investigators with crime scene reconstruction, as it did in the Harmony Montgomery case. Improved technology testing and capability characterization of body fluid analyses, supported by NIJ, can aid crime labs as they standardize crucial forensic techniques.
According to Ghemrawi, “The support from NIJ … enabled us to explore cutting-edge techniques like DNA methylation, mRNA markers, and proteomics, which are poised to revolutionize forensic biology.” She continued, “The findings from our study provide crucial data that will help forensic laboratories transition to more advanced serological methods.”
“By evaluating the error rates, sensitivity, and specificity of new serological [blood and fluid] approaches, our study paves the way for more reliable forensic investigations. The insights gained from this project are instrumental in helping forensic practitioners understand and adopt these advanced technologies,” said Ghemrawi.
About This Article
The work described in this article was supported by NIJ award number 2020-DQ-BX-0015, awarded to the Frederic Rieders Family Renaissance Foundation.
This article is based on the grantee report “Comparative Assessment of Emerging Technologies for Body Fluid Identification” (pdf, 12 pages) by Mirna Ghemrawi, Ph.D.; Jack Ballantyne, Ph.D., University of Central Florida; Bruce McCord, Ph.D., Florida International University; and Glendon Parker, Ph.D.,